immuneML is an open-source platform developed in Python for the analysis of adaptive immune receptor repertoires (AIRR). immuneML is supported by ELIXIR Norway. Read the published paper or see the preprint on biorxiv.
immuneML v3 Galaxy with immuneML v2
(deprecated from 2025) Python library Docker image


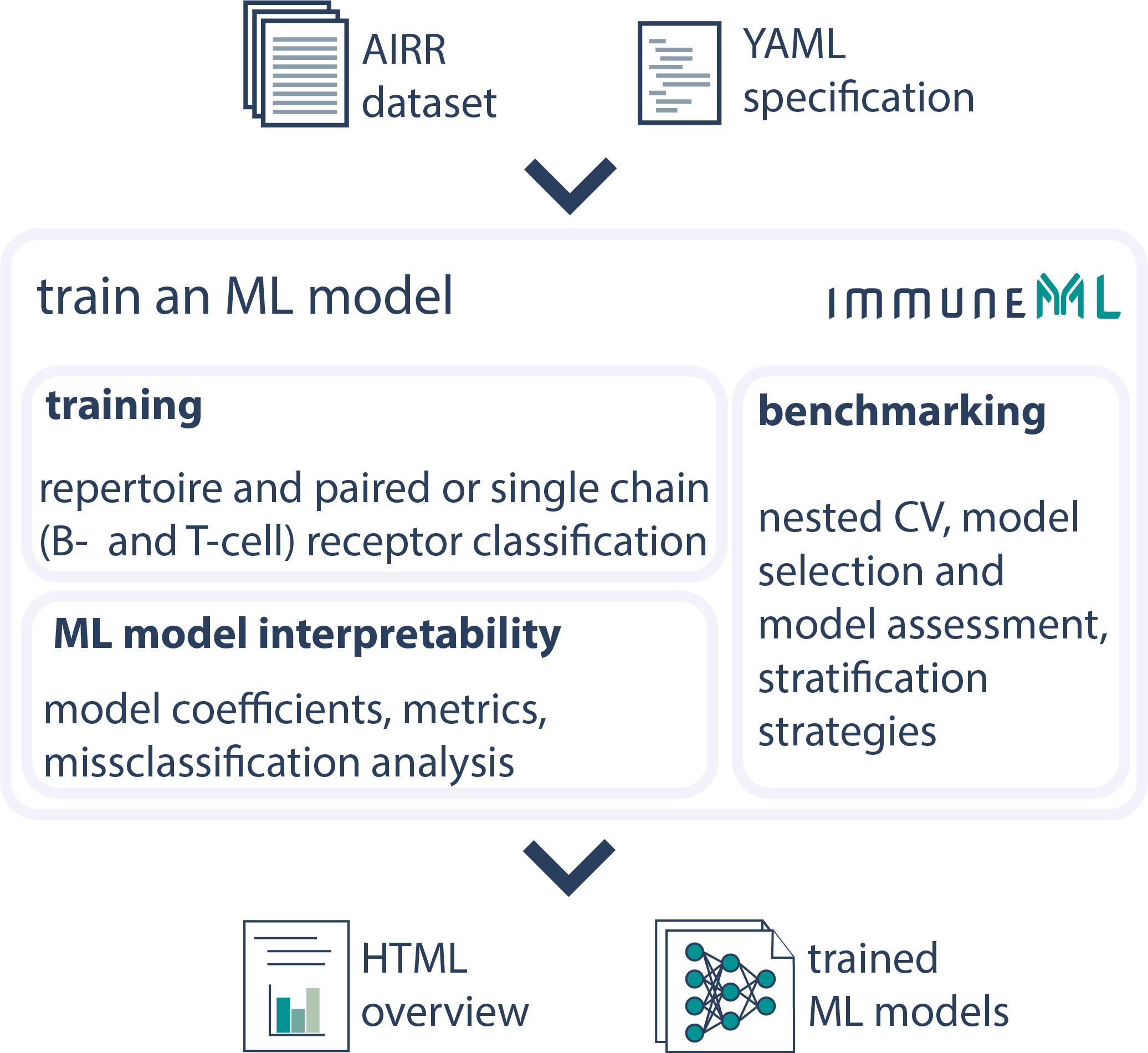
Train an AIRR Machine Learning Model
ML models can be trained for repertoire, receptor or receptor sequence classification. Multiple ML methods and encodings can be optimized over using nested cross-validation to get the optimal model. Benchmarking and interpretability analysis can be performed on the ML models to gain insights into the inner workings of the ML models.See the documentation for more details.
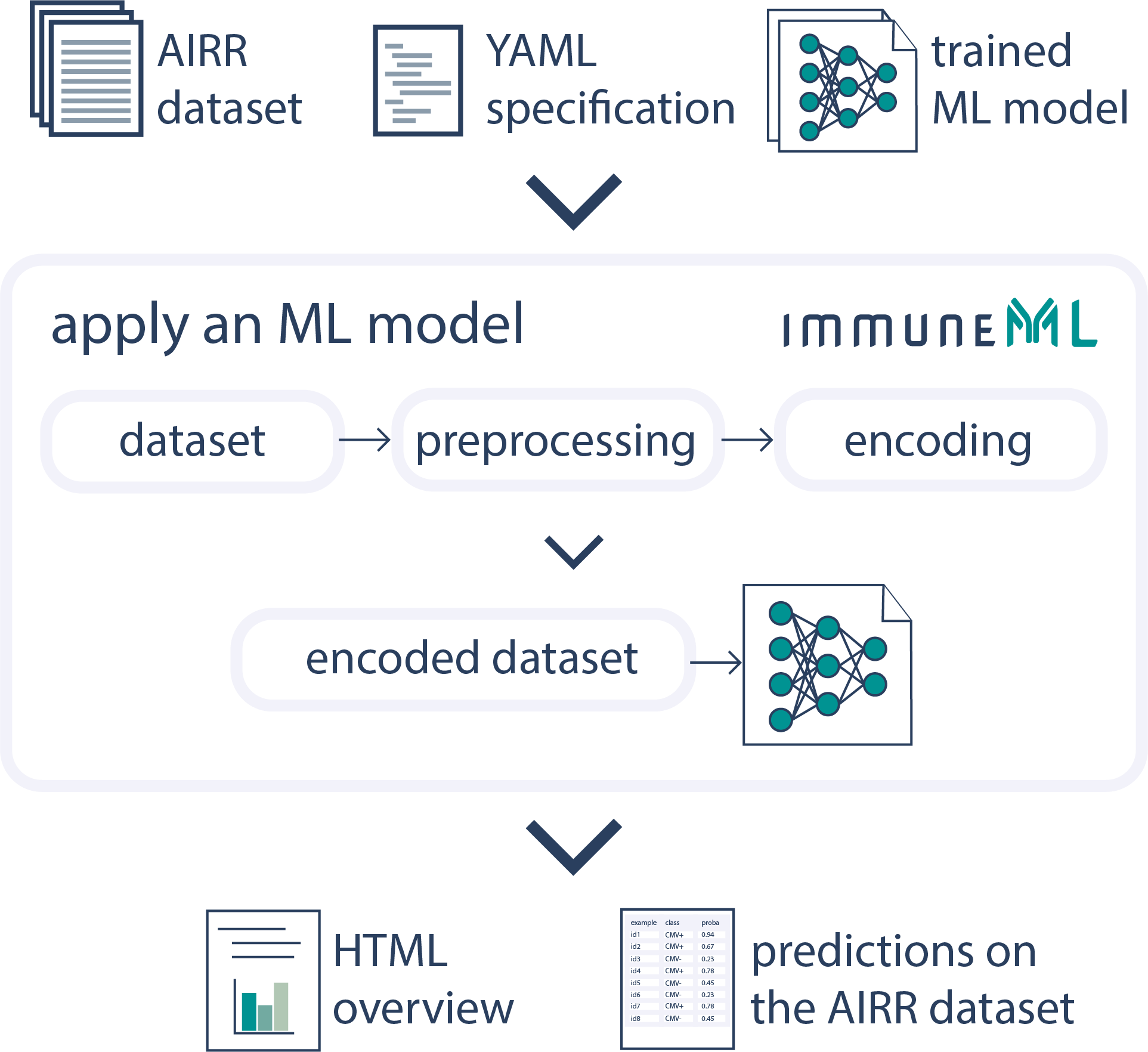
Apply a Trained AIRR Machine Learning Model
Once a machine learning model is trained, the model and encoding parameters are exported, and they can then be applied to new datasets.See the documentation for more details.
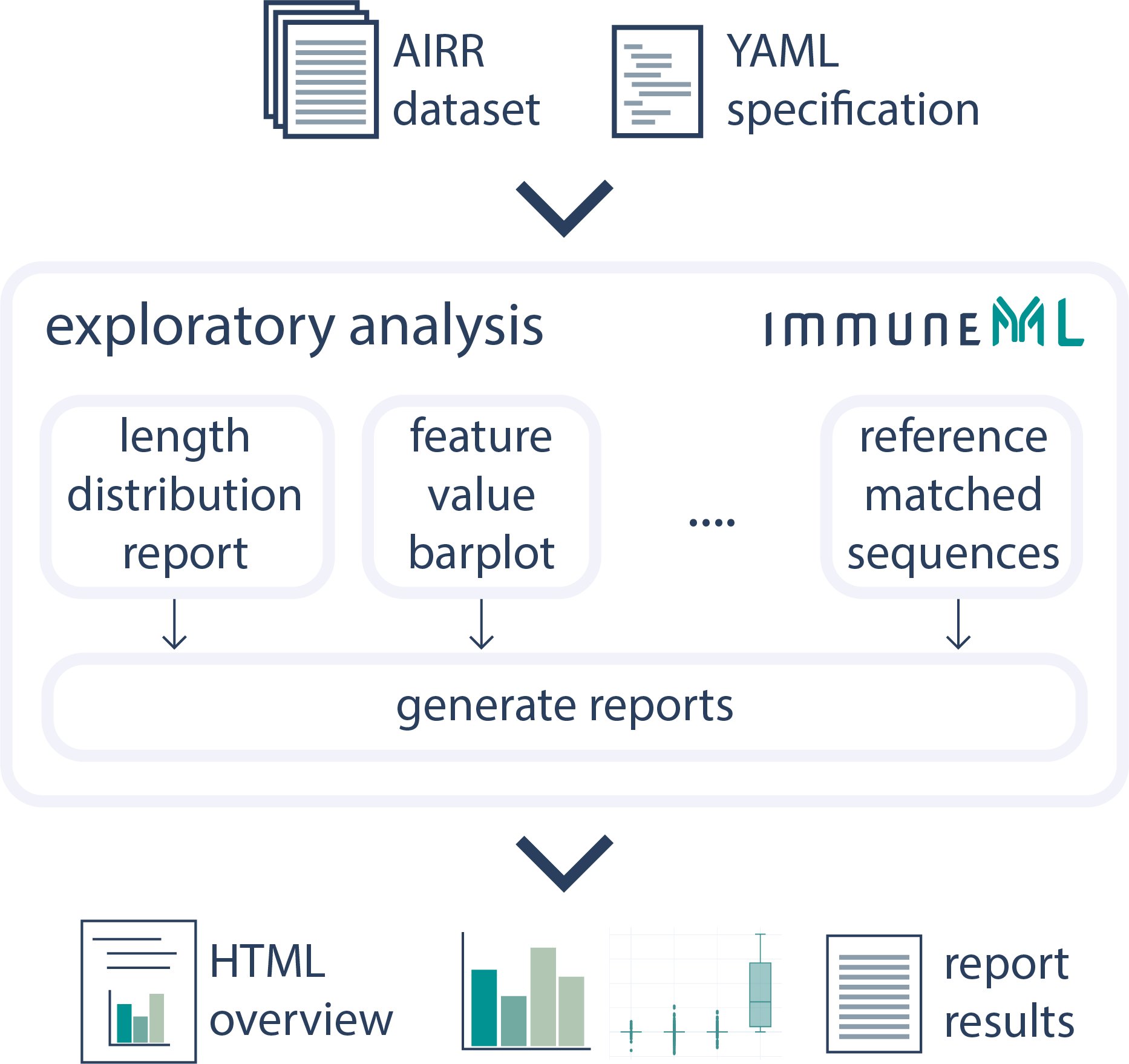
Perform Exploratory Analysis of Datasets
The Exploratory Analysis instruction can be used to explore the properties of an immune dataset before diving into training a machine learning model.See the documentation for more details.
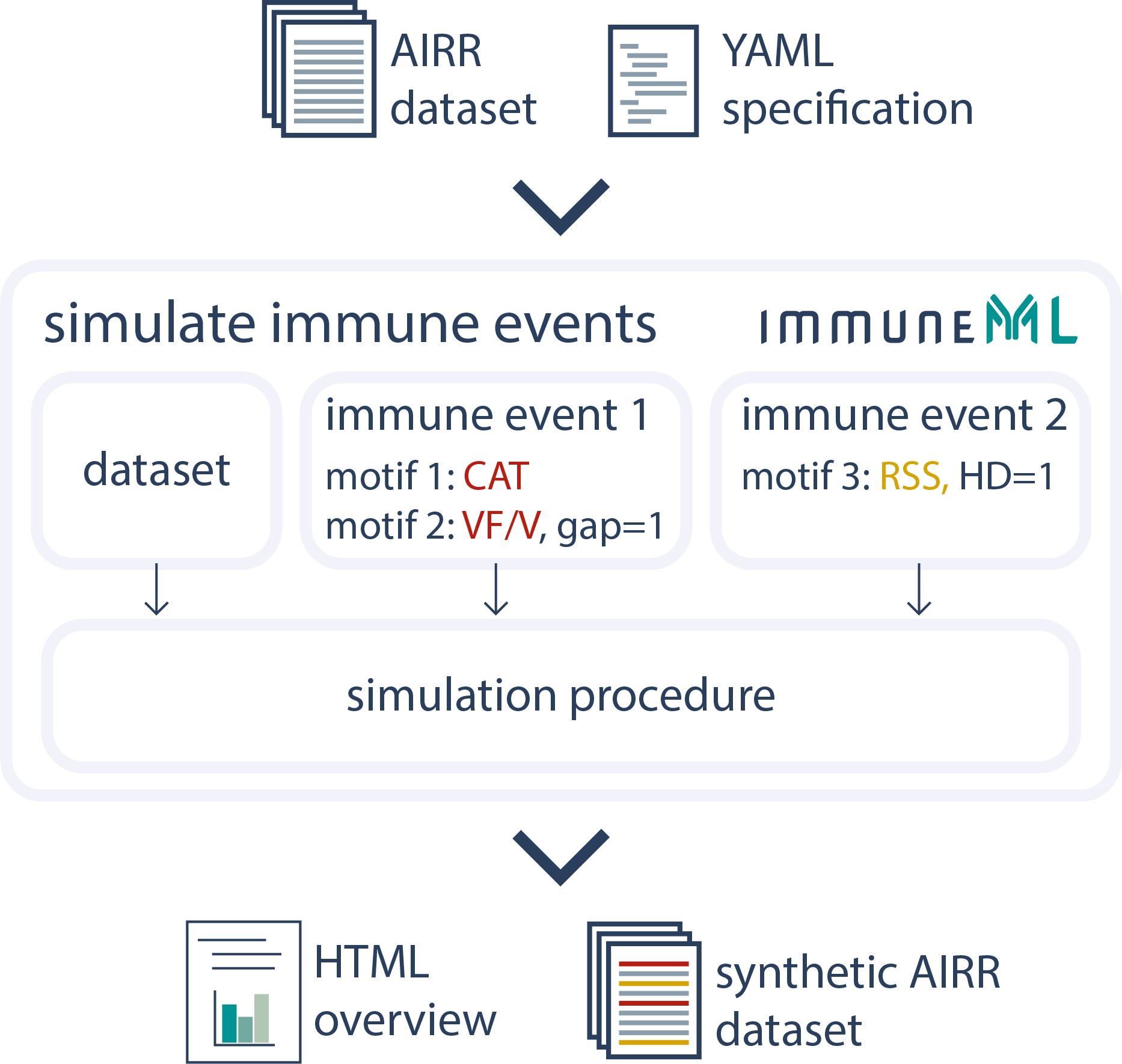
Simulate Synthetic Datasets
To simulate immune events into synthetic or into experimental datasets, immuneML offers the Simulation instruction working on both repertoire and receptor level. It supports complex implanting schemes to generate useful datasets for benchmarking.See the documentation for more details.
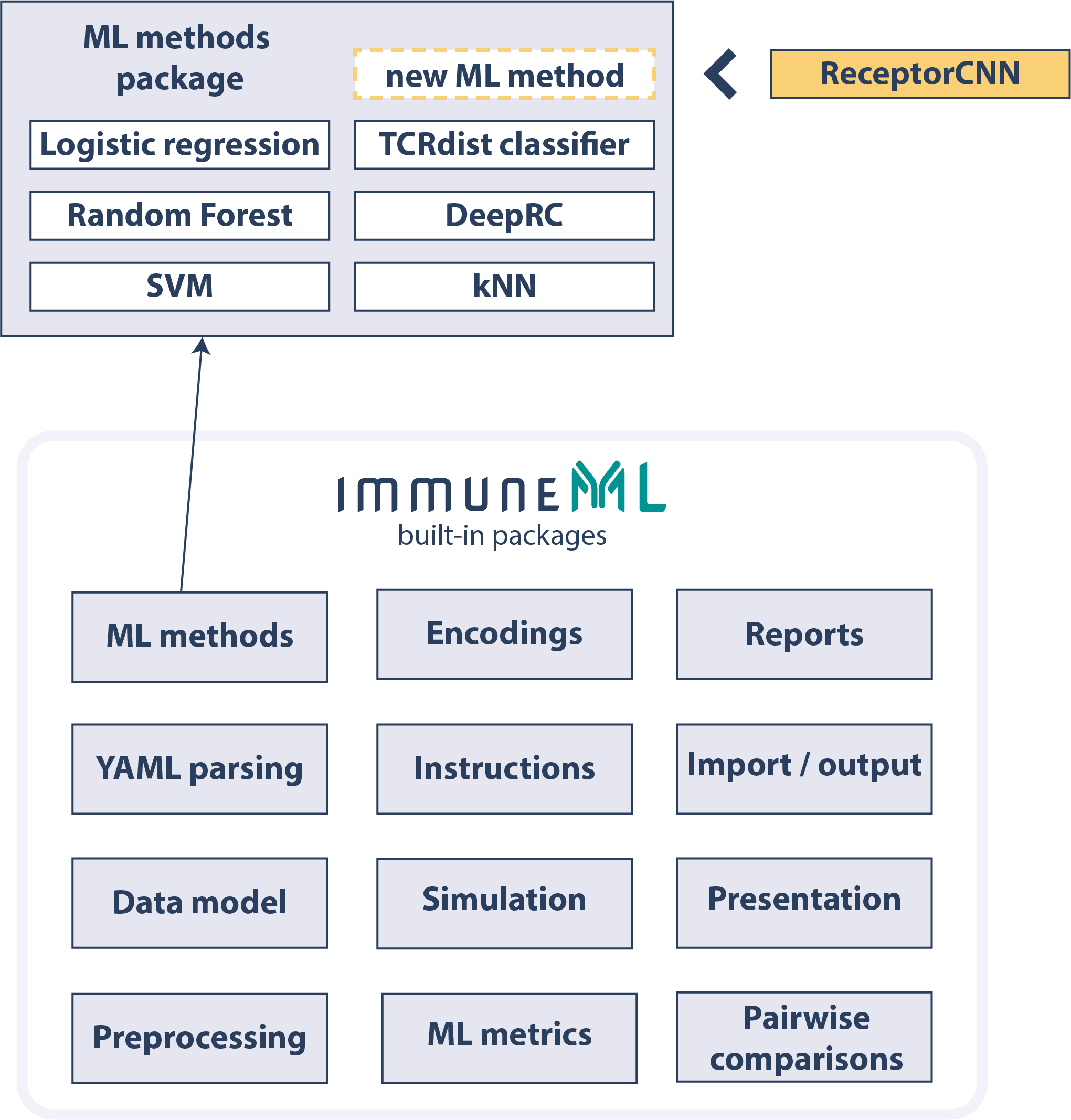
Extend The Platform With New Components
immuneML provides extensive developer documentation to encourage extending the platform with new components, such as new machine learning methods, encodings, or reports.See the developer documentation for more details.



